Improved infectious diseases research and surveillance in Ethiopia through capacity building in bioinformatics and sequencing
July - AUGUST 2019
From left to right: Prof. Gemeda Abebe, Agumas Shibabaw, Emmanuel Rivière, Abyot Meaza, Annelies Van Rie, Setegn Eshetie, Wubet Birhan, Olifan Zewdie, Melashu Balew, Getu Balay and Mulualem Tadesse.
From July 29th to August 16th, 2019, the TORCH consortium organized the second training of the Ethiopian VLIR project, together with Prof. Gemeda Abebe (Jimma University) and Dr. Conor Meehan (Institute of Tropical Medicine). For the second training, nine young Ethiopian scientists were selected:
Abyot Meaza: national TB reference laboratory, Addis Ababa
Zegeye Bonsa: Mycobacteriology Research Centre, Jimma University, Jimma
Getu Balay: Mycobacteriology Research Centre, Jimma University, Jimma
Agumas Shibabaw: University of Gondar, Gondar
Melashu Balew: Amhara Public Health Institute, Bahir Dar
Olifan Zewdie: Wollega University, Nekemte
Setegn Eshetie: University of Gondar, Gondar
Wubet Birhan: University of Gondar, Gondar
Yihun Mulugeta: Bahir Dar University, Bahir Dar
Building upon our experiences from the first training, the training sessions of the second training were organized on two different locations: from 29/7 until 9/8 in South Africa at Stellenbosch University and from 12/8 until 16/8 in Ethiopia at Jimma University.
Sample preparation & sequencing
For the second training, the DNA extraction protocol was changed to a commercial kit from Qiagen (DNEasy Ultraclean Microbial kit). The use of this kit was further optimized to increase the yield and quality of Mycobacterium tuberculosis DNA, by including an extra overnight lysozyme digest. Two protocols based on this kit were used to perform DNA extraction: one for solid microbial cultures and one for liquid cultures. During the second training we decided to work with real Mycobacterium tuberculosis cultures in the Biosafety level 3 facilities in South Africa, instead of the surrogate Mycobacterium smegmatis cultures we used for the first training. Furthermore, for the second training we decided to focus more on the DNA extraction part of the Whole Genome Sequencing wet-lab procedure, because this is the most important step and library normalization and preparation could be outsourced. Library preparation was shown to the trainees during a live demonstration from Dr. Nabila Ismaïl. DNA extractions were performed twice in the facilities in South Africa during the first two weeks of the training and again twice in the facility in Jimma, Ethiopia, during the final week of the training.
Dr. Nabila Ismaïl demonstrating the library preparation for whole genome sequencing to the trainees.
Data analysis: Linux and Galaxy
During the first two weeks of the training, several sessions were organized on data analysis. The trainees were given the same Linux tutorial that was taught during the first training. While the first training focussed on using the MTBSeq pipeline from the command line, we opted to switch to a Galaxy based software pipeline, due to its user-friendliness and low hardware requirements (since it runs on a Galaxy server). The trainees were taught how to use ‘Snippy’, a rapid haploid variant calling and core genome alignment tool, developed by Prof. Torsten Seemann. The local server access was managed by Dr. Peter Van Heusden. Additionally, the trainees were taught how to use the open-source and online available Mycobacterium tuberculosis Whole Genome Sequencing analysis tool PhyResSE.
Research projects
During the three-week course, the trainees were assigned to develop their own research project conceptually, that fits with the scope of the course and their own interests. To this end, several discussion sessions with the trainees were organized by Prof. Annelies Van Rie and Dr. Conor Meehan. During the third week of the training (in Ethiopia) Annelies Van Rie taught multiple sessions on defining a research question, specific objectives, study design and sample size estimations.
Abyot Meaza: “Tuberculosis transmission dynamics in refugee camps”
Zegeye Bonsa: "Tuberculosis permeability between refugee camps and local communities"
Wubet Birhan: “Tuberculosis clustering and super-spreaders in prisons”
Agumas Shibabaw: “Transmission of rifampicin resistant tuberculosis in the Amhara region”
Melashu Balew: “Acquired resistance during treatment of rifampicin resistant tuberculosis”
Getu Balay: "Genetic basis of cord formation of Mycobacterium tuberculosis"
Setegn Eshetie: "Tuberculosis transmission between patients and health care workers"
Olifan Zewdie: "Phylogenetic analysis of extrapulmonary tuberculosis strains"
Yihun Mulugeta: did not develop a research idea as he left the training early
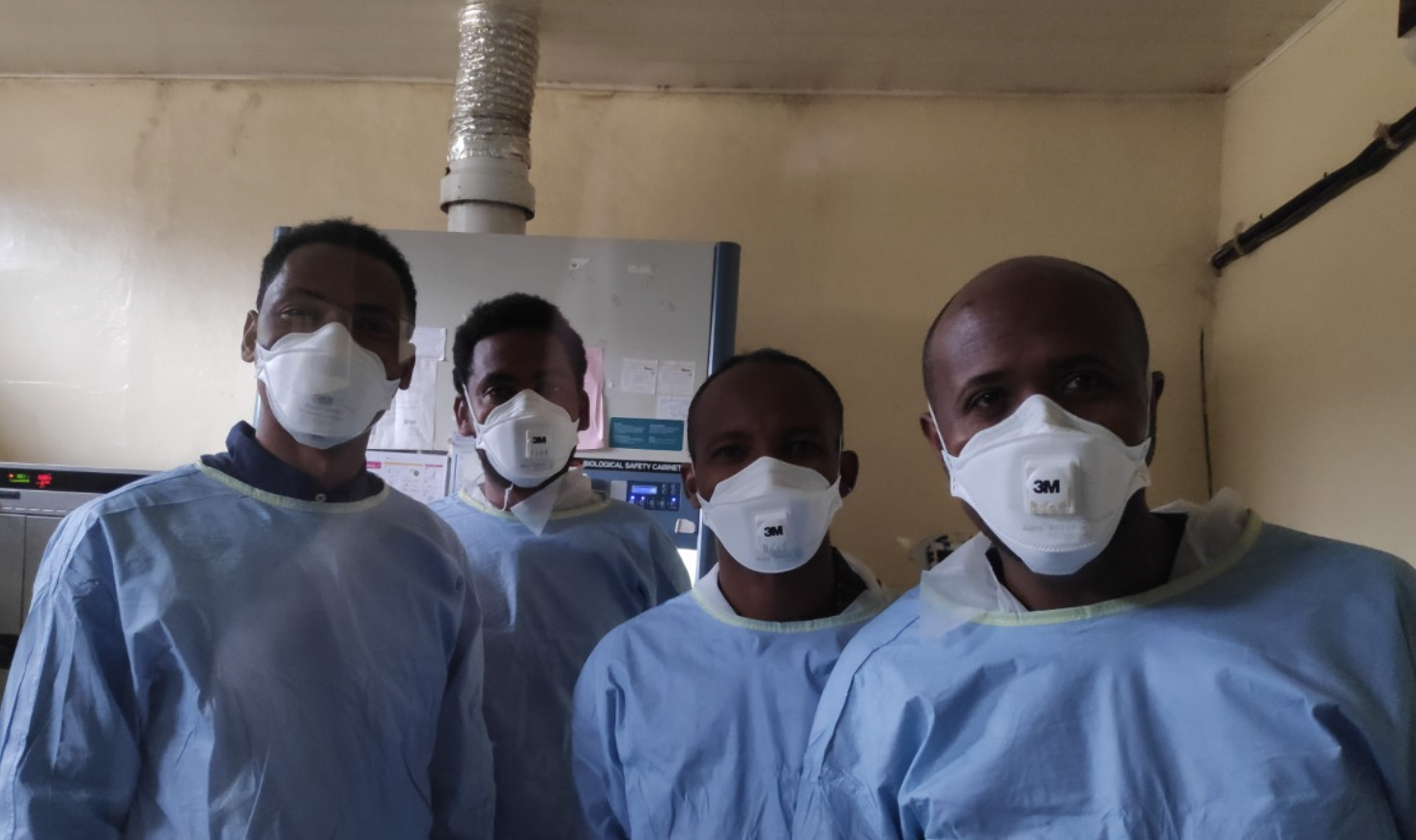
Trainees in the BSL-3 laboratory in Jimma University. From left to right: Zegeye Bonsa, Olifan Zewdie, Melashu Balew, Abyot Meaza.
Wrap up & future perspectives
The successful second VLIR-UOS training resulted in eight of nine trainees developing an interesting research concept implementing whole genome sequencing and tuberculosis. With the two trainings combined, we managed to train 15 Ethiopian researchers on whole genome sequencing for tuberculosis research. This is however not the end of the project, as we will continue to work with the trainees who want to further develop their research concepts into a pilot project.
We recently published an open-access article on our experiences organizing these training sessions and the challenges we faced: “Capacity building for whole genome sequencing of Mycobacterium tuberculosis and bioinformatics in high TB burden countries”. This article includes a systematic review to assess African research output in the field of tuberculosis as well as the training schedule we used during our trainings.
Check it out: https://doi.org/10.1093/bib/bbaa246
Back